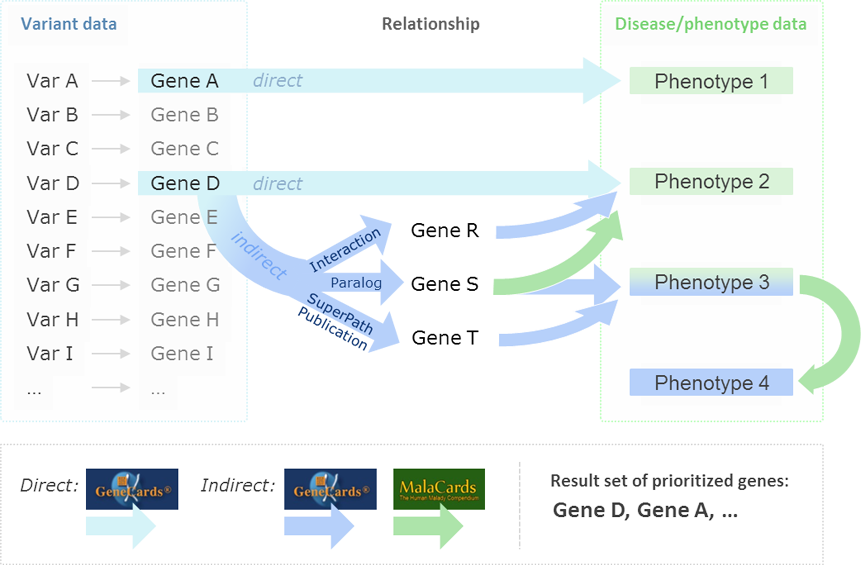
Indirect association between genes and disease are based on shared pathways, interaction networks, paralogy relations, domain-sharing, and mutual publications.
An example of an indirect GeneCards-based inference of GeneA to Phenotype PhenX is when PhenX is found to be associated with GeneB, which in turn shares a pathway with GeneA. Such gene-to-gene relationships are also formed (among others) by interaction networks, paralogy relations, domain-sharing, and mutual publications. MalaCards, in turn, allows one to produce a comprehensive phenotype search expression by using its data about diseases, underlying symptoms, and their relationships. The degree of mutual linking is quantified via endogenous search scores. VarElect provides a robust algorithm for ranking genes within a short list, and pointing out their likelihood to be related to a disease, enabling the researcher to perform the last decision step in deep sequencing runs in a fast and objective manner.
Yaron Guan Golan
Comments